Documentation
Content
Cell lines
The cell line content page provides a summary over all available cell lines at DSMZCellDive. An overview shows the cell line, cell type, and species. Furthermore, the DSMZ ACC No. is given including a link to the DSMZ catalogue, where more information is available and the cell line can be purchased. The last column indicates if the cell line is featured in any RNA-seq data panel.
Browse all cell linesSearch and filter
Cell lines can be searched by their name or DSMZ ACC No. When clicking on More filters, a popover with more filter options will appear. Here, cell lines can be filtered by species and cell types. Both fields feature auto suggestions when you start typing. Furthermore, only cell lines that belong to a certain RNA-seq panel can be selected here.
Integrated cell line data
When a single cell line is selected by clicking on its name, a cell line summary page is opened. On this page, all information on this particular cell line is shown, e.g. certain meta data, the STR profile or the COI DNA barcoding report, and RNA-seq projects.
RNA-seq summary
If the cell line is featured in one of the RNA-seq panels, charts on these data can be triggered by clicking on one of the buttons in the panel. After a short loading time, either a histogram or a bar chart with all normalised expression levels are shown.

List of genes
The genes content page lists all human genes and some meta data, e.g. the position in the human genome, database identifiers from Ensembl and Entrez and a short gene description. Gene names correspond to GENCODE names (v38). The list of genes can be filtered by gene name, description, Ensembl-ID, or Entrez-ID. The description search is always a contains-search. For gene names, one can use an asterisk as wildtile character (Example).
Browse all genesRNA-seq data
Each RNA-seq panel has its own overview page that can be reached via the sidebar or the starting page. On this page, further information on this project are given, including literature to cite when using the data.
Matrix description
Different to the description of the LL-100 paper (see below), RNA-seq data were quantified via Salmon and normalised via DESeq2. Following values are given:
- normalised values: calculated via DESeq2 within one RNA-seq project
- tpm: transcripts per million reads - a rough measure to compare between samples
- estimated counts: Salmon's estimate of counted reads per transcripts, corresponds to counted raw reads; limited comparability between samples but recommended as basis for further statistical analysis
Please note, that if all NGS projects are selected, TPM and count values are available only, since normalisation have been calculated for each project only. For specific statistical analyses please take the estimated counts of your samples of interest as starting point.
Gene selection
Genes can be selected via an intuitive selection tool. Start typing to get genes suggested based on their names and descriptions. Clicking on the × will remove the selected gene. You can clear the whole selection by clicking on clear below. For Bar charts, you can select up to 5 genes. For Heat maps, between 2 and 50 genes can be selected.
You can also paste genes into this field (only gene names supported), if the genes are seperated by spaces, linebreaks, comma, or semicolon. The gene selection is saved as a cookie for your convenience.
Example bar chart
Bar charts
Bar charts are generated using the Plotly.js library. They are interactive, one can zoom in by selecting parts of the graph, pan by tagging the axes and hide/show single data sets by clicking on the legend. You can save the chart as PNG by clicking on the that appears after hovering the plot.
Bar charts can display multiple genes (up to 5). Grouping will be done by gene or cell line, depending on user selection.
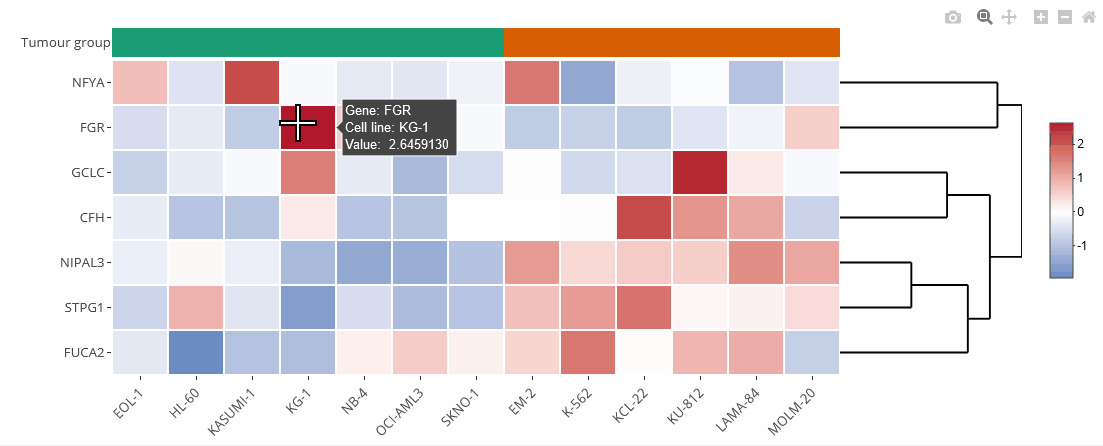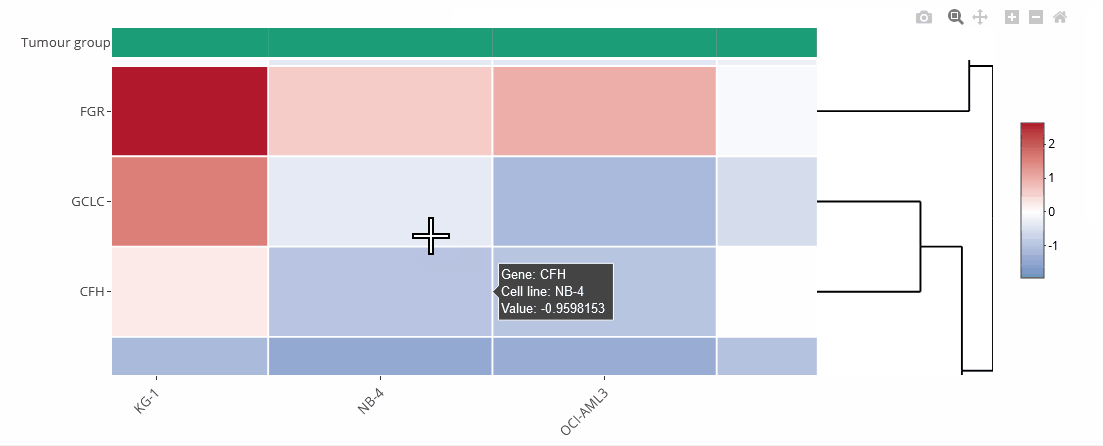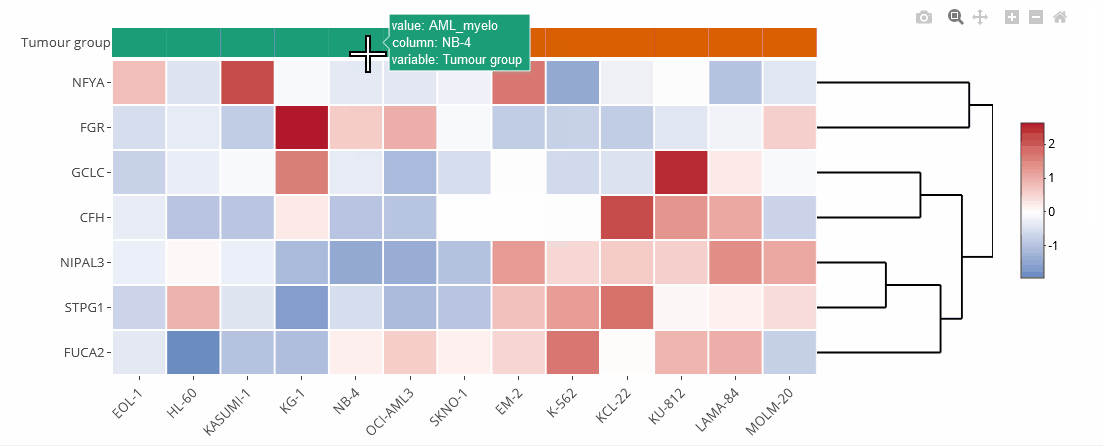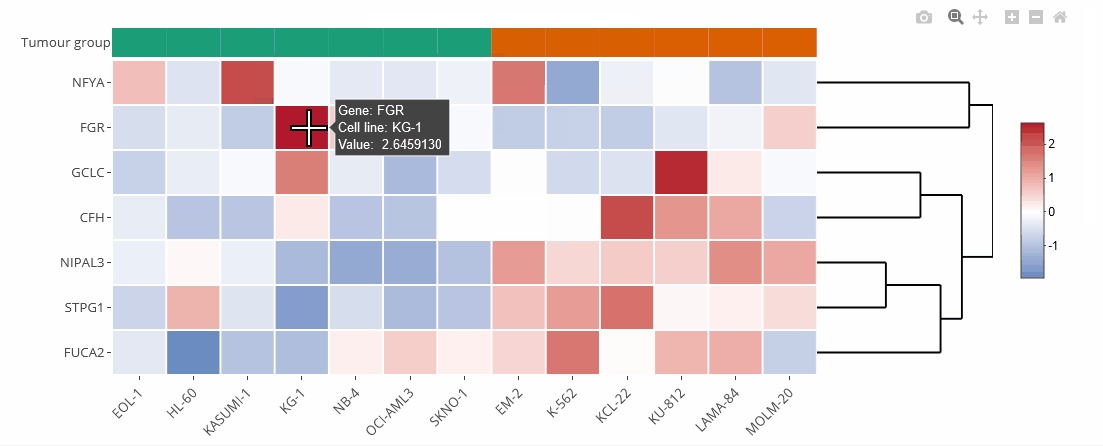
Example heat map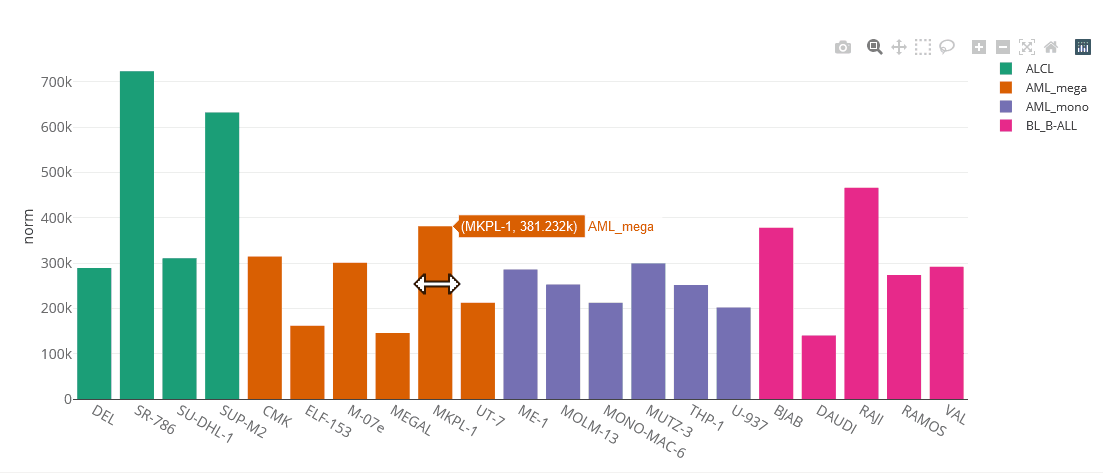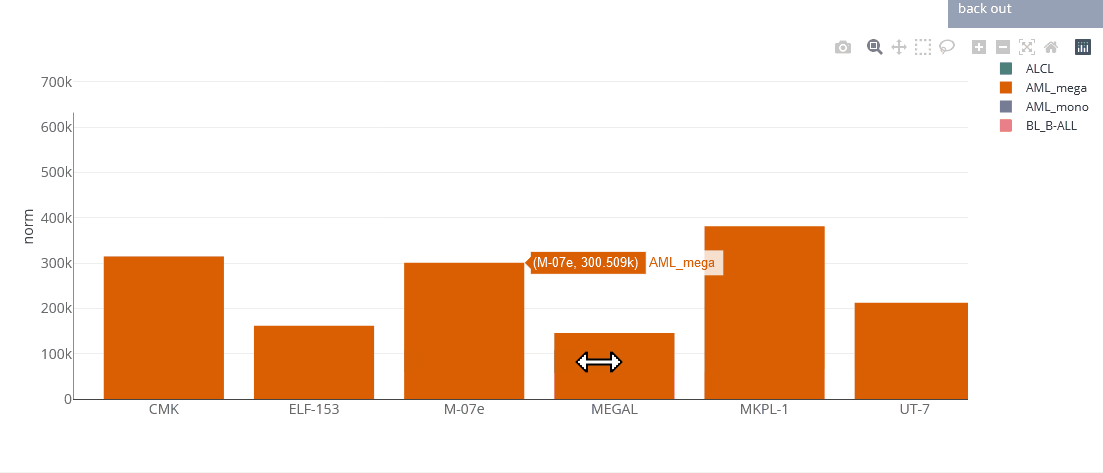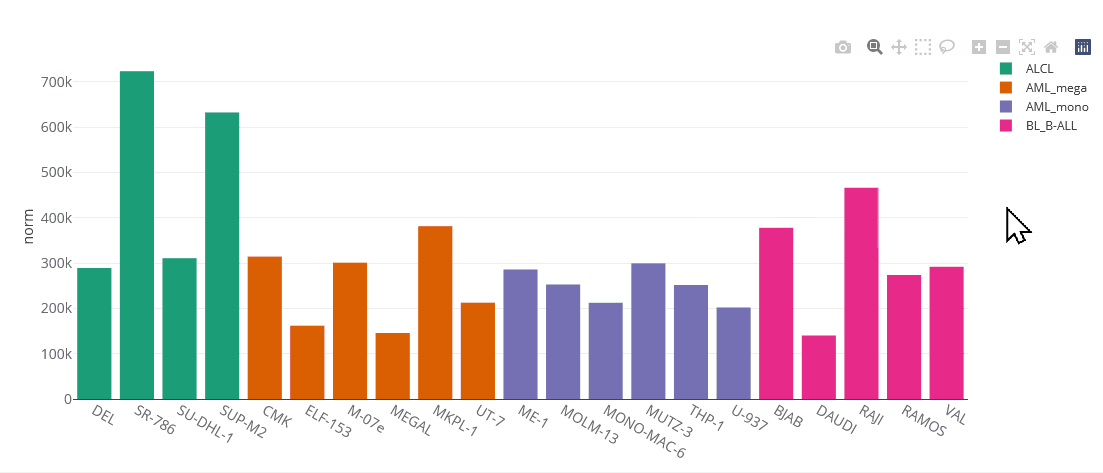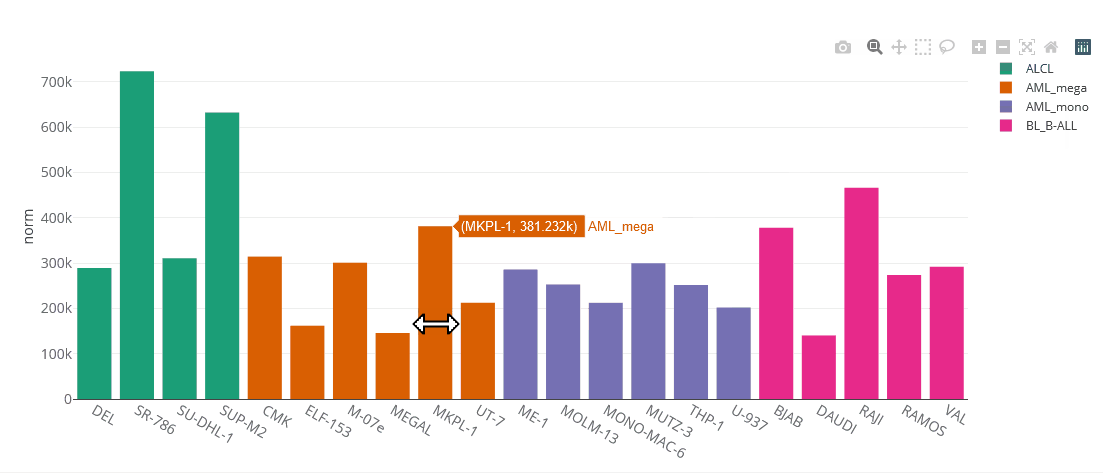
Heat maps
Heat maps are generated using the Plotly.js library and the R package heatmaply. They can be zoomed and downloaded the same way as the bar charts.
Heat maps can be clustered by genes (default) or cell lines. Clustering can also be disabled (none). The order of the genes is identical to the selection unless they are clustered. On top of the heat map is a color legend indicating the tumour entity. Hover over the labels to get more information.
Short tandem repeats for human cell lines
Short Tandem Repeats (STRs) are crucial for the authentication of human cell lines. Thus, we implemented a search engine comprising 17 STR loci and more than 4,500 data sets from cell lines of different sources.
STR search engineThe STR Search
The search tool offers a form, where one can enter each STR value manually. If more alleles are needed, they can be added using the + button. Up to six alleles are possible. Click on Example to see an example. It is also possible to paste data from e.g. Excel files. To do this, please click on Use text input and follow the instructions in the popup.
The result page contains a table with your query and the best matching STR profiles. The first column shows the similarity. Read more about this score here. The next two columns give information on the cell line and its source. The following columns contain the STR profiles. Bold red fonts indicate mismatches from your query. The result can be downloaded as CSV file that can be opened with MS Excel.
The similarity score
The similarity score is calculated as stated by Tanabe et al:

Authentic or not authentic, a cruical question: STR profiling is the recommended approach for authentication testing of human cell lines. To improve its effectiveness, however, a method by which STR can account for genetic drift arising from the passage of malignant cells is needed. Algorithms based on a set of match criteria, seventeen STR loci, and amelogenin analysis were found to successfully discriminate between related and unrelated samples. The criteria (100 to 80 % match = authentic) used here are intended to make the interpretation of STR profile search results more visible. Therefore, results highlighted in green are usually authentic as long as the cells do not carry a deficiency in DNA mismatch repair (MMR). Results highlighted in yellow indicate a distant relationship when there is a drift of STR alleles in MMR-negative cell lines. Clearly incorrect cell lines are present when the background is red.
Read more about this topic here.
COI DNA barcoding for animal cell lines
While STR data are only used for human cell lines, the mitochondrial cytochrome c oxidase I (COI) gene is widely used to identify species of animal cell lines. DSMZCellDive offers COI DNA barcoding reports for all animal cell lines in the database. You can browse through them, view them in more detail and download each report as PDF.
If you are looking for an identification engine, we recommend to use BOLDSYSTEMS.
Browse COI DNA reportsHLA typing data
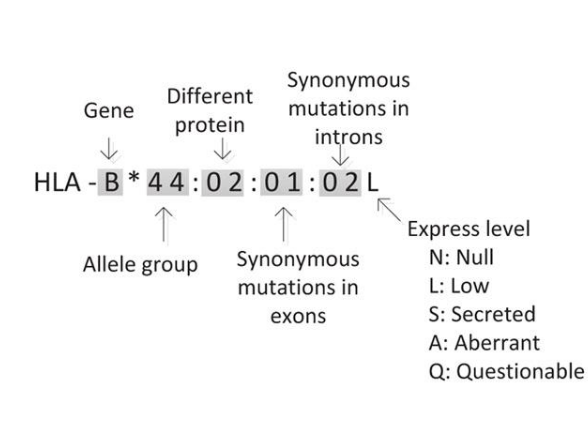
Human leukocyte antigen (HLA) genes encode proteins in the major histocompatibility complex (MHC) which play a central role in discriminating self and non-self. MHC Class I proteins (HLA-A, -B and -C) bind to and present intracellular antigens on the cell surface for cytotoxic T-cells, which trigger apoptosis if non-selfpeptides are detected. MHC Class II proteins (including HLA-DPB1, -DQB1 and -DRB1), on the other hand, present extracellular proteins to helper T-cells which mediate the adaptive immune response.
The HLA gene cluster on chromosome 6 is highly polymorphic and suitable for cell line authentication. Furthermore, HLA typing is important for cancer research and for determination of tissue compatibility, in which tumour neoantigen binding to HLA surface proteins and rejection of specific HLA alleles play a role.
Here, HLA typing was determined on RNA-seq data via arcasHLA, an alignment-based tool (Orenbuch et al, 2020).
Protein expression was not determined.
How to cite
If you found DSMZCellDive to be useful for your research, we will be happy if you cite us.
DSMZCellDive
Koblitz J., Dirks, W.G., Eberth, S., Nagel, S., Steenpass, L., & Pommerenke, C. (2022) DSMZCellDive: Diving into high-throughput cell line data. F1000Res, 11:420. DOI: 10.12688/f1000research.111175.2
The presented data
LL-100 RNA-seq data
Quentmeier, H., Pommerenke, C., Dirks, W. G., Eberth, S., Koeppel, M., MacLeod, R., Nagel, S., Steube, K., Uphoff, C. C., & Drexler, H. G. (2019). The LL-100 panel: 100 cell lines for blood cancer studies. Scientific reports, 9(1), 8218. DOI: 10.1038/s41598-019-44491-x
Breast Cancer RNA-seq data
Pommerenke C, Nagel S, Haake J, Koelz AL, Christgen M, Steenpass L, Eberth S. Molecular Characterization and Subtyping of Breast Cancer Cell Lines Provide Novel Insights into Cancer Relevant Genes. Cells. 2024; 13(4):301. DOI: 10.3390/cells13040301
STR profiling data
Dirks, W. G., MacLeod, R. A., Nakamura, Y., Kohara, A., Reid, Y., Milch, H., Drexler, H. G., & Mizusawa, H. (2010). Cell line cross-contamination initiative: an interactive reference database of STR profiles covering common cancer cell lines. International journal of cancer, 126(1), 303–304. DOI: 10.1002/ijc.24999
STR authentication and similarity score
Capes-Davis, A., Reid, Y. A., Kline, M. C., Storts, D. R., Strauss, E., Dirks, W. G., Drexler, H. G., MacLeod, R. A., Sykes, G., Kohara, A., Nakamura, Y., Elmore, E., Nims, R. W., Alston-Roberts, C., Barallon, R., Los, G. V., Nardone, R. M., Price, P. J., Steuer, A., Thomson, J., … Kerrigan, L. (2013). Match criteria for human cell line authentication: where do we draw the line?. International journal of cancer, 132(11), 2510–2519. DOI: 10.1002/ijc.27931

